deaths / year — ≈ 647 per day by meningitis
Current tools avoid only part of the problem
Resistance of S. pneumoniae has significantly increased
Harnessing ML & Quantum to Fight Meningitis

CS and Cyber-Security
Passionate about cybersecurity and ethical hacking, Ishaan brings innovative solutions to protect digital infrastructures and ensure data security in our interconnected world.

CS and Biomed Engineering
Bridging the gap between technology and healthcare, Ishita focuses on developing cutting-edge medical technologies and bioengineering solutions to improve patient care.

CS and Quantum Info Science
Exploring the frontiers of quantum computing and information science, Aditya works on revolutionary quantum algorithms and their practical applications.
deaths / year — ≈ 647 per day by meningitis
Resistance of S. pneumoniae has significantly increased
Bacteria like those causing meningitis are becoming resistant, and current drugs can’t keep up. Drug discovery today is too slow and expensive.
Qure AI uses AI to design new molecules and quantum computing to confirm their stability, creating faster, smarter antibiotics.
Cleaned the original databases using python scripts with pandas to parse and remove empty or unknown data and taking only relevant data, all the data was converted to CSV files with proper headings and clear distinctions. Then the CIF files were parsed and converted to CSV's with all their positional data.
We built an SVM model to generate potential meningitis cures, analyzing four bacterial causes. Using features like MIC, IZ, molecular weight, and Lipinski’s Rule of 5, we identified novel, drug-like molecules with promising antibacterial activity.
Implemented the Variational Quantum Eigensolver by finding ground state energy. Used Active Space Reduction to minimize problem set. VQE was used to validate and compare the ground state energies of the existing molecule and the AI-generated molecule.
threshold = median(Standard Value)
for each molecule:
if Standard Value <= threshold:
label = effective (1)
else:
label = ineffective (0)
for up to max_mutations:
randomly choose mutation_type ∈ { add_atom, replace_atom }
if add_atom:
pick random_atom ∈ { C, N, O, F, Cl }
connect it to random_position in molecule
if replace_atom:
pick random_position in molecule
pick new_atom ∈ { C, N, O, S }
replace atom at that position
Molecular Weight = random normal(mean, std of dataset)
AlogP = random normal(mean, std of dataset)
RO5 Violations = random choice from dataset values
Standard Type = random choice from dataset types
Standard Value = random choice from dataset values of that type
x = transform_row(new_molecule)
probability = model.predict_proba(x)
if probability > 0.6:
accept as "Generated Candidate"
else:
discard
This project trains Support Vector Machines (SVMs) on bacterial drug datasets by converting molecular and experimental properties into numeric features. Molecules are labeled effective or ineffective based on median activity values, enabling the SVM to learn patterns that distinguish promising drug candidates.
Using RDKit, the system mutates existing SMILES strings, randomly sampling molecular properties to generate realistic new compounds. Each candidate is evaluated by the trained SVM, and only those with predicted effectiveness above 60% are retained. The pipeline outputs novel drug-like molecules, storing results per bacterium for further analysis.
The Variational Quantum Eigensolver (VQE) algorithm is used to minimize a cost function. In this project, we are using the VQE to find the ground state energy. We first find the Hamiltonian. This represents the energy of the system. Then we choose an appropriate ansatz. The ansatz is a guess that is formed by a parameterized circuit. We used Qiskit's EfficientSU2 function.
The classical optimizer optimizes the circuit after every run. The parameters of the ansatz is adjusted and the circuit is run again. This adjustment happens after every iteration to minimize the cost function. We do 10 iterations in an attempt to find the ground state energy.
PROGRAM VQE
INPUT: csv_file_path (molecular coordinates)
OUTPUT: convergence_plot, energy_data
BEGIN
// Initialize parameters
max_iterations:10
ansatz_repetitions: 2
energy_history: empty_list
// Step 1: Generate Hamiltonian
PRINT "Generating molecular Hamiltonian..."
molecular_data: READ_CSV(csv_file_path)
hamiltonian: generate_hamiltonian(molecular_data)
num_qubits: calculate_qubits_needed(molecular_data)
num_electrons: count_active_electrons(molecular_data)
// Step 2: Create Quantum Ansatz
PRINT "Creating quantum ansatz..."
ansatz: create_ansatz(num_qubits, num_electrons, ansatz_repetitions)
initial_parameters: random_parameters(ansatz.num_parameters)
// Step 3: VQE Optimization Loop
PRINT "Running VQE optimization..."
current_parameters: initial_parameters
FOR iteration = 1 TO max_iterations DO
// Evaluate energy expectation value
quantum_state: ansatz.execute(current_parameters)
energy: expectation_value(hamiltonian, quantum_state)
energy_history.append(energy)
// Classical optimization step
gradient: compute_gradient(energy, current_parameters)
current_parameters: update_parameters(current_parameters, gradient)
PRINT "Iteration", iteration, "Energy:", energy
END FOR
// Results processing
optimal_energy: minimum(energy_history)
execution_time: current_time() - start_time
molecular_formula: create_formula(molecular_data)
// Generate outputs
PRINT "Saving results..."
SAVE_CSV(energy_history, "energy_convergence.csv")
CREATE_PLOT BEGIN
x_axis: [1, 2, ..., length(energy_history)]
PLOT_LINE(x_axis, energy_history)
ADD_HORIZONTAL_LINE(optimal_energy)
SET_LABELS("Function Evaluation", "Energy (Hartree)")
SET_TITLE("VQE Energy Convergence for " + molecular_formula)
SAVE_PLOT("vqe_results.png")
END CREATE_PLOT
// Final summary
PRINT "Simulation Complete!"
PRINT "Molecular formula:", molecular_formula
PRINT "Ground state energy:", optimal_energy, "Hartree"
PRINT "Execution time:", execution_time, "seconds"
PRINT "Total evaluations:", length(energy_history)
END
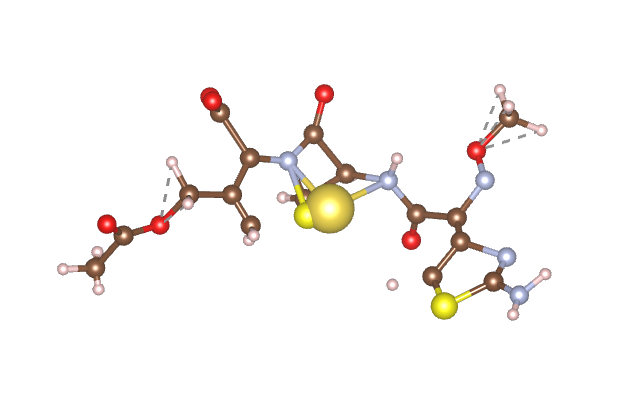
We implemented the VQE algorithm by using active site reduction methods to find the ground state energy of the molecules. The quantum algorithm used a maximum of 10 iterations and approximately 10-20 qubits for a simulation. We simulated Cefotaxime sodium, a molecule that is effective in targeting Streptococcus pneumoniae. The generated molecule for targeting Streptococcus pneumoniae achieved a significantly lower ground state energy than the existing drug. This shows that the machine learning model is generating highly stable drugs.